Findings Knowledge Base¶
What is the Findings Knowledge Base? (KB)¶
The Findings Knowledge Base (KB) is one of the most important features in Canopy. It helps you reuse content across all of your reports, so you don’t have to write the same findings all the time. You can also link finding KBs and tools (e.g. Nessus) together to automatically overwrite tool findings during the import process. This allows you to replace vendor content with your content to automate the tailoring of content write-up for your team and your clients.
Using the findings KB and building it up over time can help:
Reduce time spent on reporting, as you no longer have to write similar findings every time.
Ensure a higher degree of quality in your write-ups by using an approved write-up.
How does it work?¶
The findings KB is a repository of your write-ups. You can re-use them when adding your findings to the test phases. When you add a KB finding to a phase, you can modify it as required - this becomes a copy of the KB finding, allowing for full adaptation to your specific engagement.
We do not currently maintain a curated list of finding write-ups, as most of our users have their own content.
Using the findings KB¶
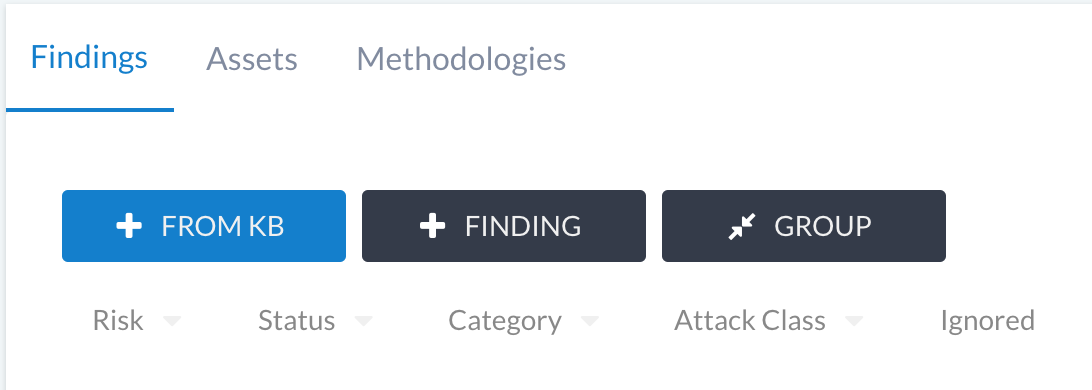
Adding KB findings to your phase can occur via two approaches:
The + FROM KB button on the findings list.
Auto-add using the tool importers.
You can also use the findings KB when grouping other findings together.
Adding KB findings to a phase¶
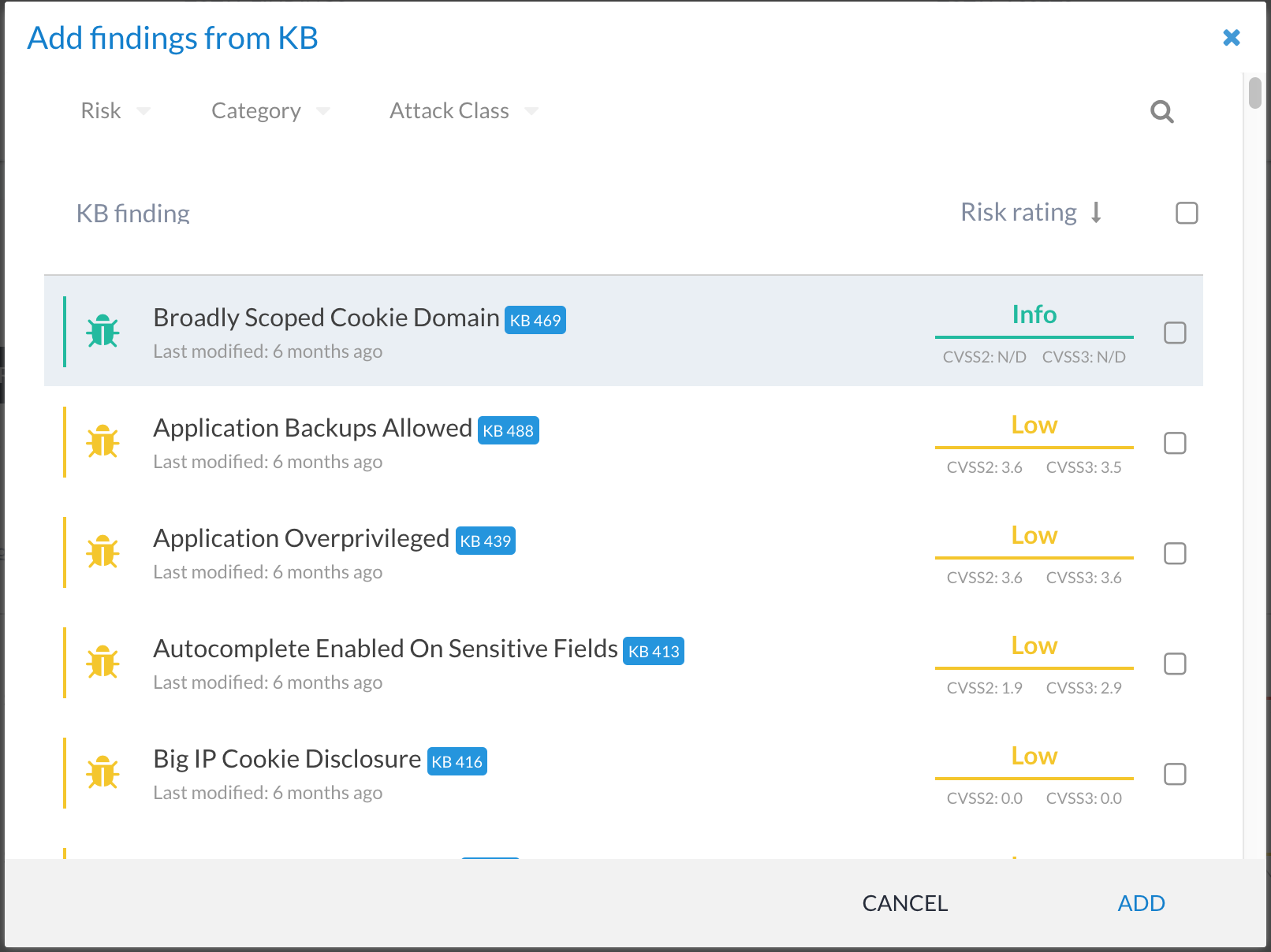
You can add findings to a phase by clicking on the + FROM KB button below:
This presents you with a list of all of the approved KB entries. These can then be selected and added to the phase:
KB findings and tool imports¶
In order to use your own KB when importing from tools, the linking to the tools must be set up first. See the KB finding and tool linking section below on how this is set up.
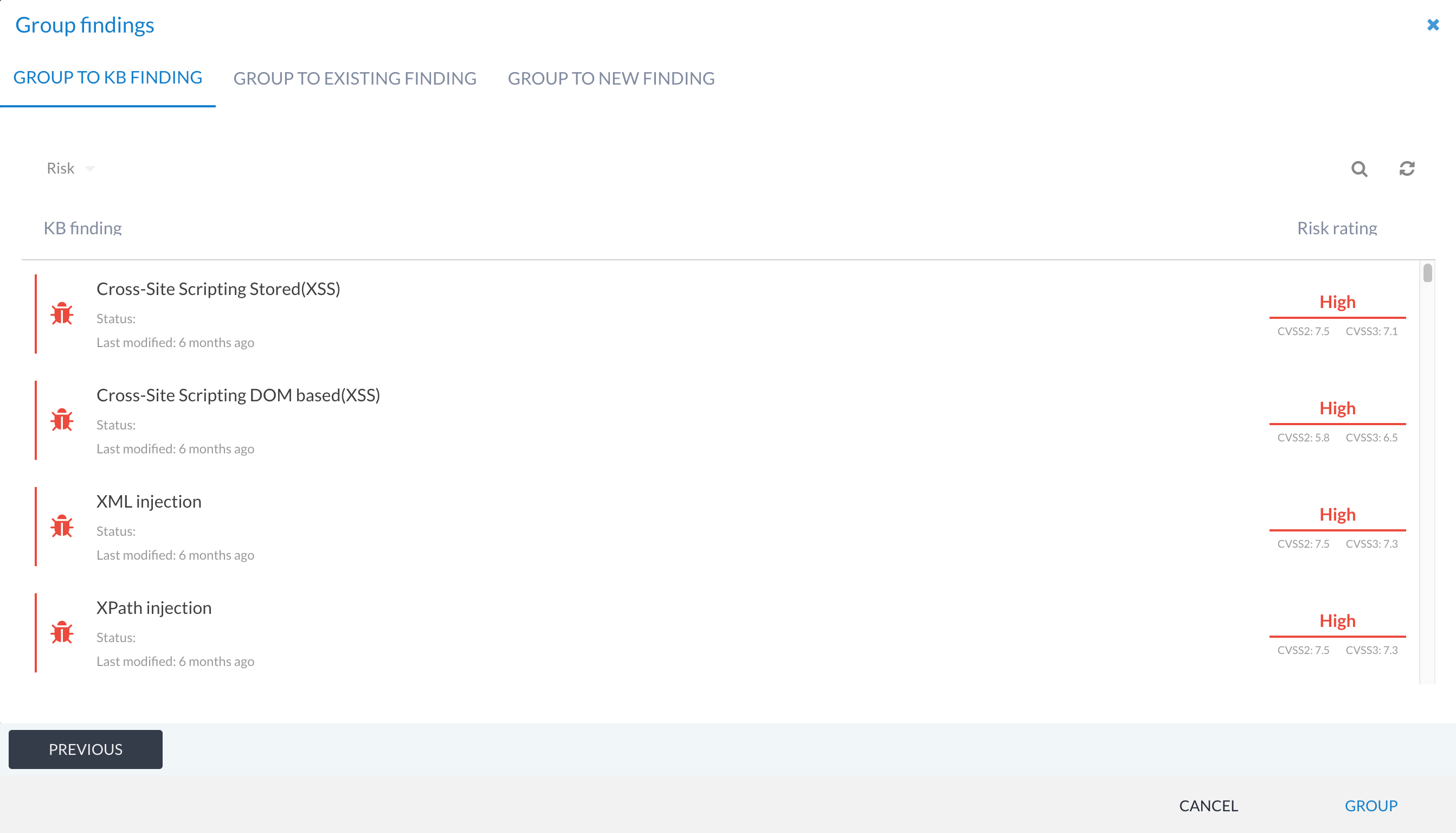
Grouping to KB findings¶
When you use the GROUP option on a findings list, one of the options presented on the second screen of the grouping workflow is to group the selected findings into a KB finding. This is useful when you want to reuse a write-up, but haven’t yet linked it to any tool data, or you just want to group a manual finding to a KB finding write-up instead.
Managing KB findings¶
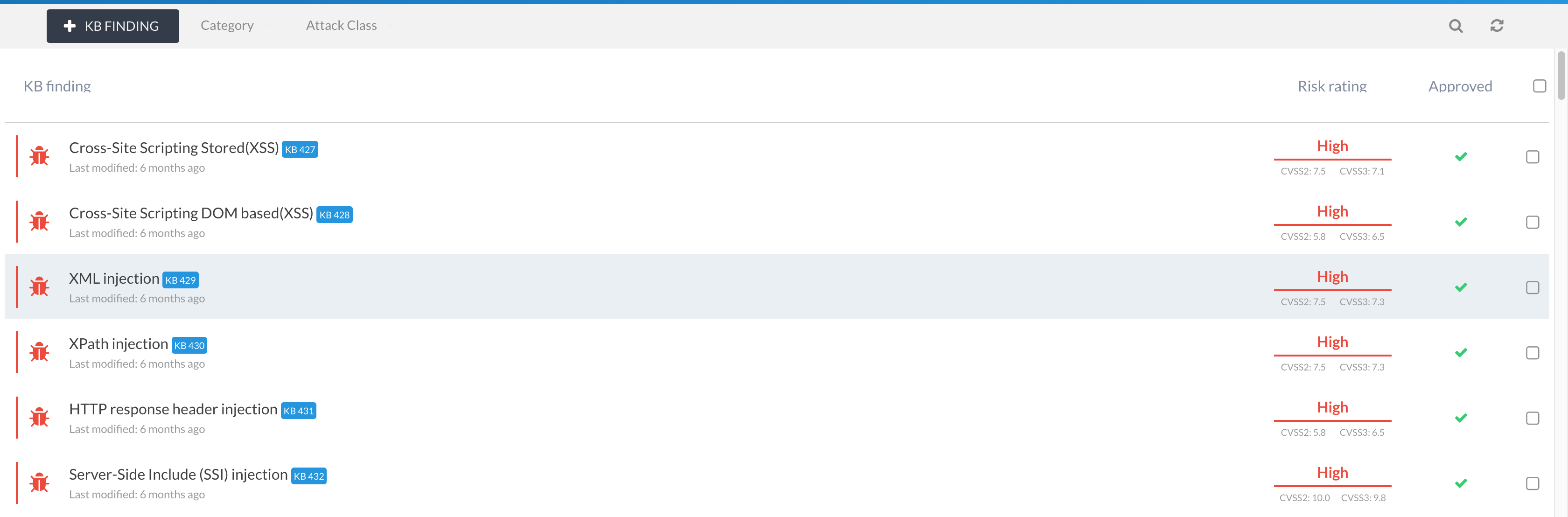
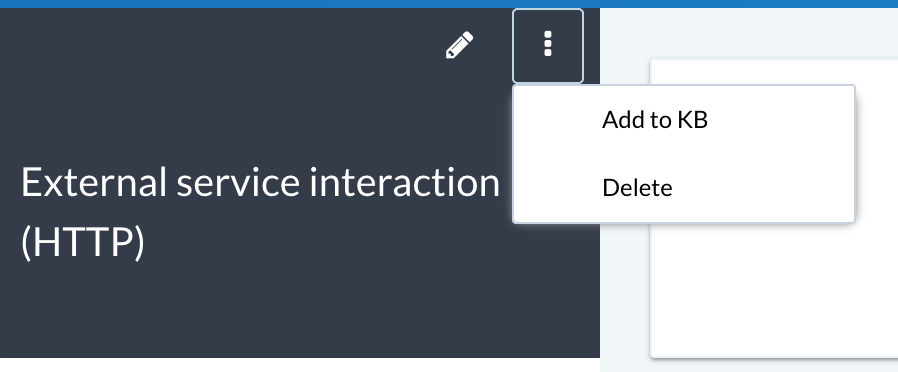
You can add new KB findings either from the KB management interface (Knowledge Base (KB) from the main navigation menu) or from the finding view via the Add to KB menu item:
If you have KB Admin privileges, then you can add and manage KB findings from the Knowledge Base (KB) main menu item.
All users have read access to the KB.
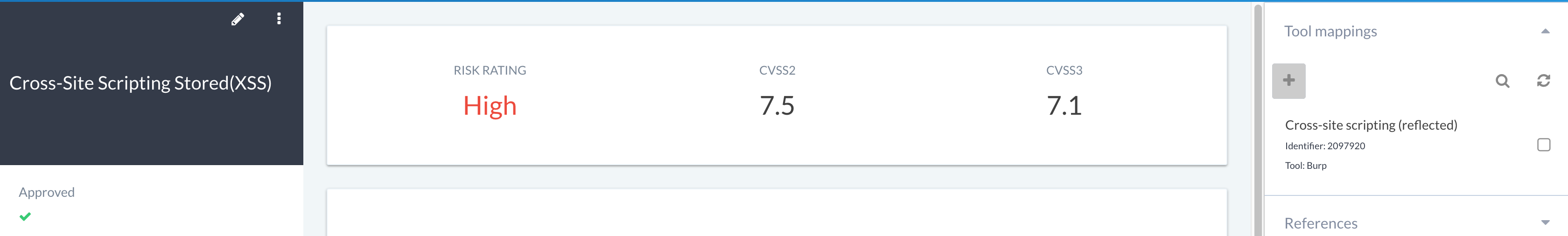
KB finding and tool linking¶
Linking KB findings with tools is a really efficient way of automatically processing tool data into your own customised write-ups, or grouping like issues together. This can be managed on a per-finding basis and accessed via the Tool mappings section in the KB finding view:
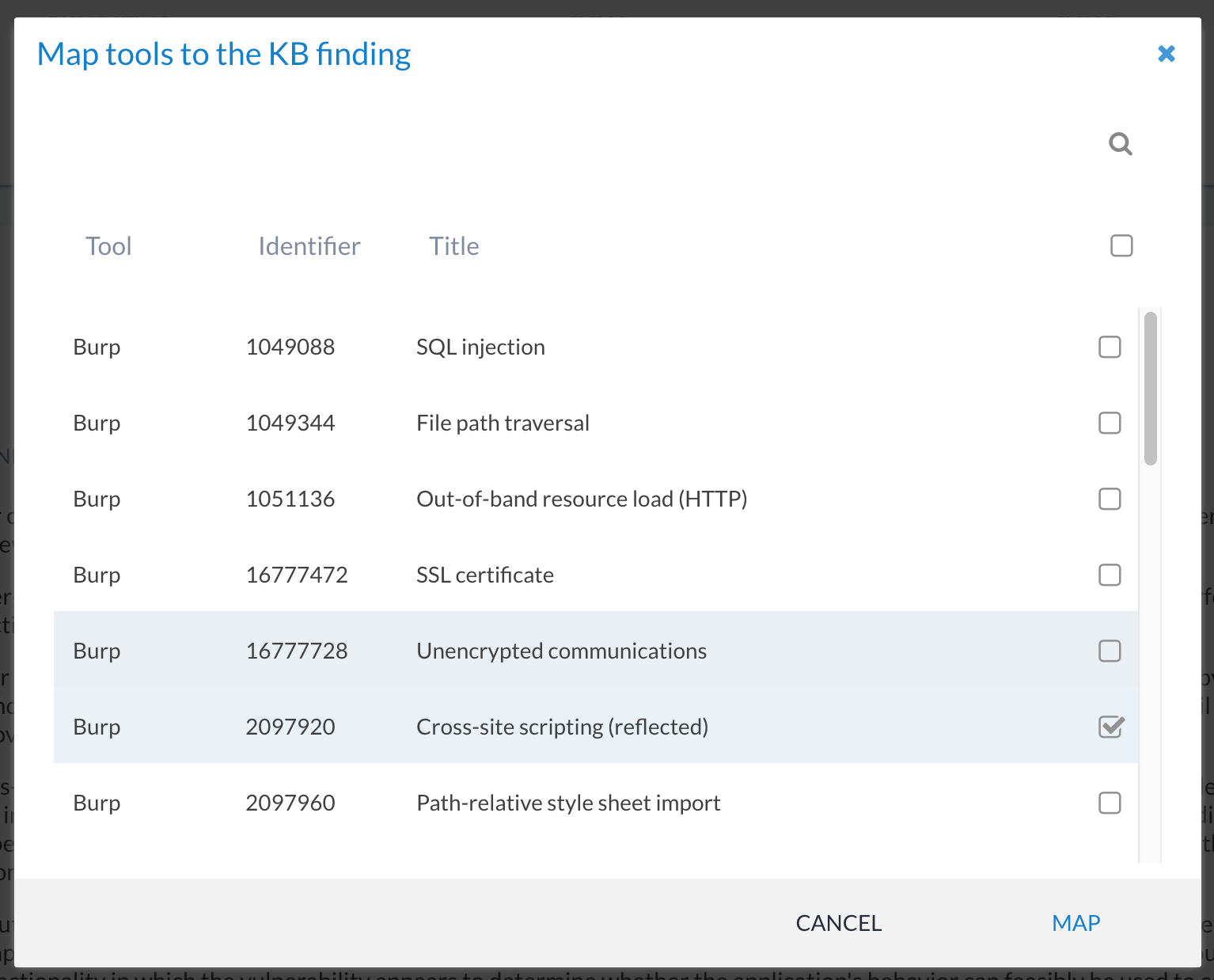
You can link one or more tools to the same KB finding. This could be useful for merging the same finding reported by different tools (e.g. SQL injection from Netsparker and from Burp Suite). Or you might want to use it to group multiple findings that are similar (e.g. all Oracle missing patches or findings relating to browser security headers). This is quite a powerful feature of Canopy and can be used to greatly speed up projects that depend on tool-generated data.
To add new finding-tool mappings, click on the + button and select the tool identifiers (i.e. the ID for the tool check) to associate:
If you add a link incorrectly, you can delete them from the KB finding view.
Importing an existing KB¶
There are several approaches to importing an existing KB:
Using the standard XLSX structure (see
this spreadsheet).Writing a custom importer management plugin.
Standard XLSX importer¶
The sample XLSX structure provides a list
of the default fields that can be added to the KB. You can also add
additional fields relating to the findings model. For example, if you
added a custom field called additional_information (see
Custom fields [TODO]), you would simply add a new column to
the XLSX and ensure that the header cell has the field name (not the
description) set.
To import, run the following command:
usage: canopy-manage kb_import [-h] [--version] [-v {0,1,2,3}]
[--settings SETTINGS] [--pythonpath PYTHONPATH]
[--traceback] [--no-color] [--delete]
[--pretend]
INPUTXLSX
canopy-manage kb_import my_kb.xlsx
Custom KB importer¶
If you have data in a pre-defined structure that you cannot easily convert to the standard XLSX format used, then writing a custom importer may be the best approach. The goal of the custom importer is to transform your source data into Canopy’s data structure.
The following importer is provided as an example:
__version__ = '0.0.1'
import json
import os
import re
import sys
from decimal import Decimal
from textwrap import dedent
import xmltodict
from django.core.management.base import BaseCommand, CommandError
from canopy.libs.cvss2 import Cvss as Cvss2
from canopy.libs.cvss3 import Cvss as Cvss3
from canopy.libs.markdown import markdown2html
from canopy.libs.toolformats.parsers import FindingField, Parser, ToolField
from canopy.libs.toolformats.parsers.errors import UnrecognizedFormat
from canopy.libs.utils import listify
from canopy.modules.common.models import AttackClass, Category, Rating
from canopy.modules.phases.models import FindingCustomField
from canopy.modules.templates.models import (
TemplateFinding, TemplateTaxonomyItem)
from canopy.modules.templates.kbimport import import_kb
class Command(BaseCommand):
help = 'Import KB items from Sample XML file.'
def add_arguments(self, parser):
parser.add_argument(
'INPUTXML', help='XML file containing KB items to import')
parser.add_argument(
'--delete', '-D', action='store_true',
help='Delete existing KB items before import')
parser.add_argument(
'--pretend', '-p', action='store_true',
help='Show parsed KB items only; the database is left untouched')
parser.add_argument(
'--approve', '-A', action='store_true',
help='Approve all findings after import')
def handle(self, *args, **options):
xmlfname = options['INPUTXML']
delete = options['delete']
pretend = options['pretend']
approve = options['approve']
if not os.path.isfile(xmlfname):
raise CommandError('Invalid file name: %r' % xmlfname)
self._check_custom_fields()
if pretend:
findings = SampleXmlKbParser(xmlfname).parse().get('findings')
items = [i for i in findings if i]
print json.dumps(items, indent=2, default=repr)
else:
if delete:
TemplateFinding.objects.all().delete()
ids = import_kb(xmlfname, SampleXmlKbParser)
if approve:
TemplateFinding.objects.all().update(approved=True)
self.stdout.write('Imported %d KB items' % len(ids))
if options['verbosity'] > 1:
self.stdout.write('IDs: %s', ' '.join(ids))
# Verifies that any custom fields required have been created in Canopy
# before importing.
def _check_custom_fields(self):
"""Ensure that required custom fields exist.
* custom_details: Rich Text
"""
custom_fields = {
f.name: f for f in FindingCustomField.enabled_objects.all()}
expected = {
'custom_details'
}
missing = [f for f in expected if f not in custom_fields]
if missing:
raise CommandError(
'Missing custom fields: %s' % (', '.join(missing)))
# ---===[ Parser code below ]===---
def cvss2_score(cvss2):
try:
return Decimal(cvss2['basescore'])
except Exception:
return None
def cvss2_vector(cvss2):
vector = cvss2['vector']
if not vector or vector in ('NA', ''):
return None
if vector.startswith('(') and vector.endswith(')'):
vector = vector[1:-1]
try:
return Cvss2(vector).to_vector()
except Exception:
return None
def cvss3_score(cvss3):
try:
return Decimal(cvss3['basescore'])
except Exception:
return None
def cvss3_vector(cvss3):
vector = cvss3['vector']
if not vector or vector in ('NA', ''):
return None
if vector.startswith('(') and vector.endswith(')'):
vector = vector[1:-1]
if not vector.startswith('CVSS:3.0/'):
vector = 'CVSS:3.0/%s' % (vector,)
try:
return Cvss3(vector).to_vector()
except Exception:
return None
# A lookup function to map the custom 'vuln class' field to a category in Canopy.
def lookup_category(vulnclass):
if not vulnclass:
return None
try:
return Category.objects.get(type=vulnclass)
except Category.DoesNotExist:
pass
attack_classes = list(
AttackClass.objects.prefetch_related('category')
.filter(type=vulnclass))
if attack_classes:
return attack_classes[0].category
sys.stderr.write(
'Warning: Unable to find category for vuln class: %r\n' % (vulnclass,))
# mark-down to HTML conversion
def markdown(txt):
if not isinstance(txt, basestring):
return txt
return markdown2html(strip_spaces(txt))
# Custom function to map custom rating labels to Canopy's rating system
def risk_rating(risk):
rating = risk['default']
rating_map = {
'H': 'High',
'M': 'Medium',
'L': 'Low',
'I': 'Info',
}
if rating not in rating_map:
return None
return Rating.objects.get(type=rating_map[rating])
def strip_spaces(s):
if s is None:
return None
if isinstance(s, list):
return [strip_spaces(l) for l in s]
if not isinstance(s, basestring):
if isinstance(s, dict) and '#text' in s:
return strip_spaces(s['#text'])
return strip_spaces(str(s))
return dedent(s.lstrip('\n').rstrip())
class SampleXmlKbParser(Parser):
TOOL_NAME = 'Sample XML KB Importer'
XML_ROOT_TAG = 'knowledgebase'
# Note: this is where Canopy fields are mapped to the fields being passed by
# the importer. Custom functions (e.g. fn=cvss3_vector) can be used to handle
# processing/massaging of custom data.
FIELDS = [
FindingField('id', ToolField('id', fn=remove_prefix)),
FindingField('title', ToolField('vulnerability', fn=strip_spaces)),
FindingField('category', ToolField('vulnclass', fn=lookup_category)),
FindingField('background', ToolField('impact', fn=markdown)),
FindingField('description', ToolField('details', fn=markdown)),
FindingField('recommendation', ToolField('remedy', fn=markdown)),
FindingField('rating', ToolField('risk', fn=risk_rating)),
FindingField('custom_details', ToolField('custom_details', fn=markdown)),
FindingField('cvss2_score', ToolField('cvssv2', fn=cvss2_score)),
FindingField('cvss2_vector', ToolField('cvssv2', fn=cvss2_vector)),
FindingField('cvss3_score', ToolField('cvssv3', fn=cvss3_score)),
FindingField('cvss3_vector', ToolField('cvssv3', fn=cvss3_vector)),
]
def __init__(self, filename, withraw=False):
# Change default of `withdraw` to False
super(SampleXmlKbParser, self).__init__(filename, withraw=False)
def enum_findings(self, infile):
return listify(self.parse_file_as_xml(infile)['record'])
def parse_file_as_xml(self, xmlfile):
"""Override that keeps `xmltodict` from stripping leading
whitespace, since it interferes with indentation detection.
"""
try:
xml = xmltodict.parse(
xmlfile, dict_constructor=dict, strip_whitespace=False)
if self.XML_ROOT_TAG:
xml = xml[self.XML_ROOT_TAG]
except Exception:
raise UnrecognizedFormat(self.filename)
return xml